Simultaneous Screening of Multiple Repeat Expansion Disorders
Opportunity
Over 50 genetic disorders are caused by expansion of simple sequence repeats dispersed throughout the human genome, including fragile X syndrome (FXS), fragile XE non-syndromic intellectual disability (FRAXE NSID) and the spinocerebellar ataxias.
Although trinucleotide repeats are often the main cause of repeat expansion disorders, there are also other repeat expansions that are responsible for these disorders. Additionally, significantly overlapping clinical phenotypes make identifying repeat expansion disorders by signs and symptoms challenging. Therefore, symptomatic individuals have to undergo molecular genetic testing to identify causative mutations and ascertain disease status.
Currently, repeat expansion mutations are detected either through applying the standard polymerase chain reaction (PCR) together with the Southern blot analysis or the triplet-primed PCR (TP-PCR). These methods are costly, effortful and time intensive-especially when testing involves many genes.
In contrast to existing strategies, this novel method offers an assay design that simultaneously screens a patient for presence of repeat expansion mutation in two or more suspected genes-offering a more rapid and cost effective simultaneous detection of common repeat expansion.
Technology
This technology describes a single-tube assay to screen multiple genetic loci responsible for different repeat expansion disorders that are caused by expansion of the same type of repeat.
Utilising the multiplex TP-PCR method, the assay uses differentially labelled locus-specific flanking primers and a common triplet-primed (TP) primer, followed by a single capillary electrophoresis run, to detect expansion mutations involving trinucleotide repeats that are present in different genes.
Amplified products at any locus showing repeat size within the pathogenic size range, or exceeding the largest normal repeat size, can be quickly identified by capillary electrophoresis. More importantly, this single amplification reaction provides a reliable one-step mutation screening of multiple disease genes-considerably shortening the diagnostic odyssey for affected individuals.
This method can be applied to simultaneous screening of any group of disease genes which share the same repetitive sequence or its reverse complement. One example is the development of a multiplex TP-PCR assay to detect seven common spinocerebellar ataxias caused by expansion of a CAG repeat within their respective genes. Another example is the development of a duplex TP-PCR assay that allows simultaneous screening of FXS, associated disorders and FRAXE NSID by targeting the CGG and CCG repeats present at the FRAXA and FRAXE fragile sites respectively.
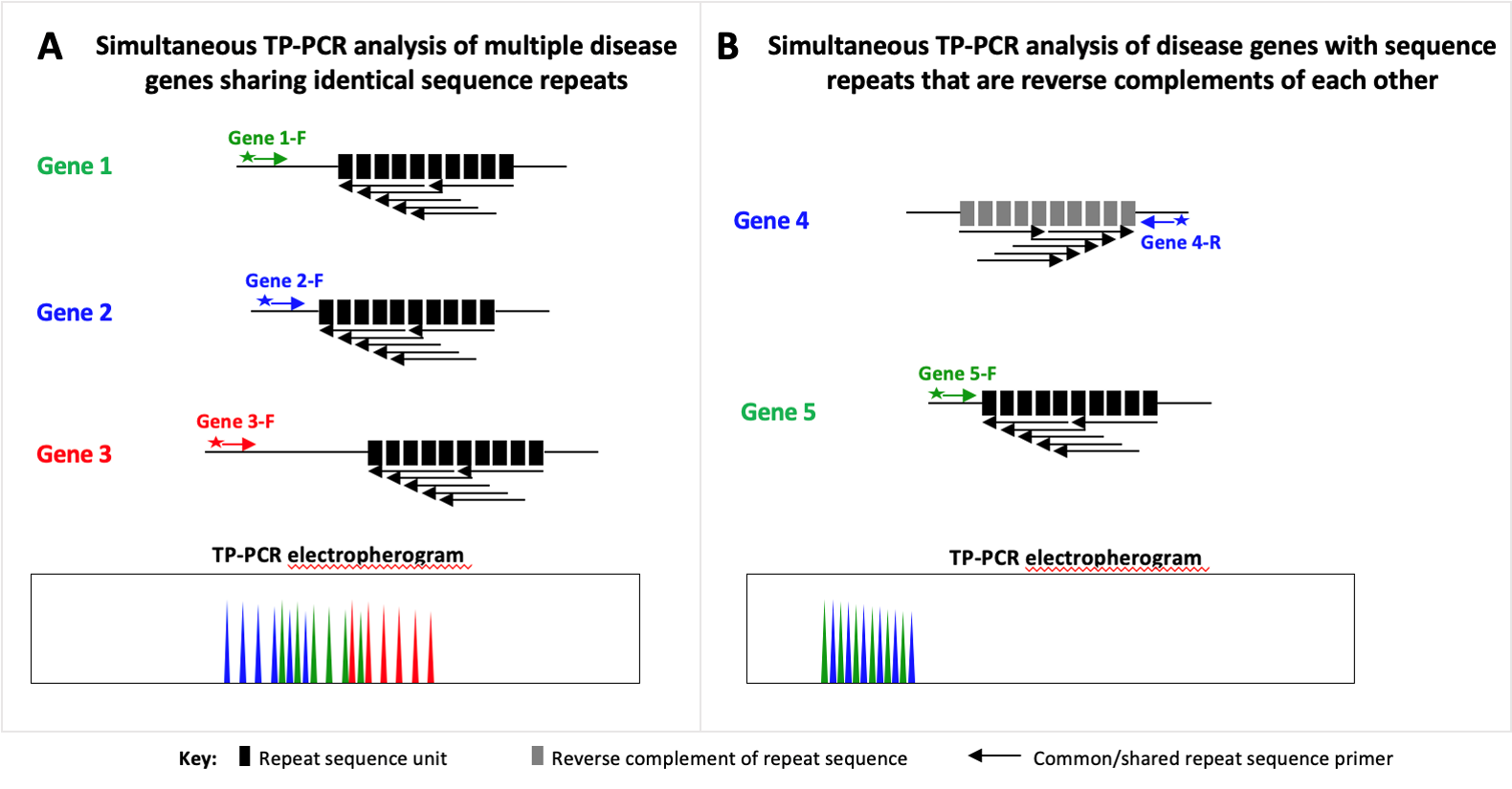
Schematic representations demonstrating simultaneous TP-PCR analysis of multiple disease genes: (A) sharing identical sequence repeats; and (B) with sequence repeats that are reverse complements of each other.


